Instrument scientist Dr Eleanor Campbell reports that an international team of researchers led by the University of Bristol (UK) have identified a possible cause of SARS-CoV-2’s increased infectivity compared to SARS-CoV (the virus which emerged in China in 2003) , which could provide a target for developing COVID-19 therapies.
Australian collaborators included researchers from the Institute of Molecular Bioscience at the University of Queensland, who sent the samples to the Australian Synchrotron.
The report, published in Science late last year, focused on the interactions of the SARS-CoV-2 spike protein and a human cell surface protein, the Neuropilin-1 (NRP1) receptor.
The SARS-CoV-2 virus is decorated with spike proteins, which are responsible for bypassing human cell membranes and entering host cells. Spike proteins do this by interacting with parts of the surface of human cells.
However, the spike protein in its natural form is not effective at adhering to host cells; it must first be cleaved into two components, and the virus uses human proteins to do this cleaving.

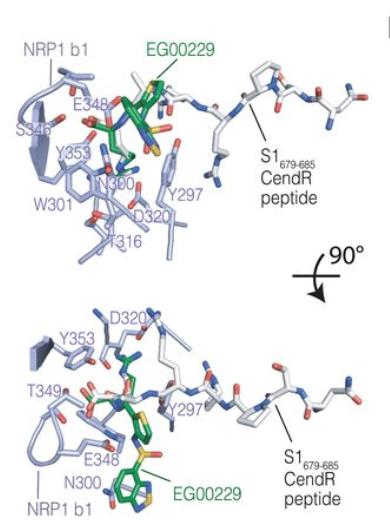
NRP1 b1–S1 CendR peptide complex superimposed with NRP1 b1–EG00229 inhibitor complex in Neuropilin-1 is a host factor for SARS-CoV-2 infection From Figure 2: Science 10.1126/science.abd3072 Shared according to Creative Commons Attribution License https://creativecommons.org/licenses/by/4.0/
After the spike protein is cleaved, one half (S1) presents an amino acid sequence that is known to interact with NRP1 on the surface of the cell.
Researchers used the MX1 beamline at the Australian Synchrotron to obtain a structure of NRP1 in a complex which included the relevant part of the S1 protein. This allowed them to determine those amino acids in NRP1 that were interacting directly with the viral protein.
The work also demonstrated that an existing small molecule inhibitor could reduce the infectivity of SARS-CoV-2.
This molecule acted as an ‘antagonist’, binding to NRP1 at the same site where the cleaved spike protein would normally bind. Thus, with the molecule in the way, interaction between the viral protein and NRP1 was inhibited.
Although it is not clear how this interaction makes SARS-CoV-2 more infective, NRP1 is overproduced in the lungs of some patients with COVID-19, suggesting the clinical relevance of this protein-protein interaction.
The interaction may not directly prevent the virus from attaching to the host cell surface, but does have an impact on infection, and is a notable point of difference between SARS-CoV and SARS-CoV-2.
Other international collaborators included the University of Tartu (Estonia), University of Zurich (Switzerland), Biological Research Centre (Hungary), University of Finland, EFTH Zurich (Switzerland), University of Liverpool, Agency for Science, Technology & Research (Singapore), and Nagoya University (Japan)


