Researchers from the University of Queensland (UQ) have used the Australian Synchrotron and cryo-electron microscopy in China to determine the three-dimensional structure of a complex enzyme found in plants microbes that could be used to develop advanced herbicides and treatments for infection.
A large international team led by Prof Luke Guddat of UQ published the structure of the enzyme acetohydroxyacid synthase (AHAS) in the journal Nature and also explained the first step in how the enzyme regulates the biosynthesis of three essential amino acids, leucine, valine and isoleucine.
“The way that the complex regulates this pathway had been unknown until now. We were finally able to explain it by understanding how the entire structure was assembled,” said Prof Guddat, who has been researching this enzyme for twenty years.

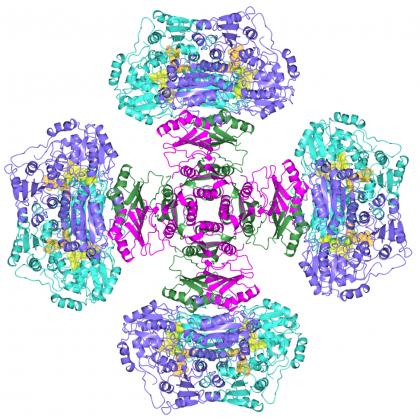
The 3D structure resembles a 'Maltese Cross'
One of the biggest surprises in the investigation was the striking configuration of the complex, which resembled the shape of a ‘Maltese Cross’.
The cross-like structure is made from regulatory subunits which form a core upon which catalytic subunit dimers (two bound proteins) are attached.
“We never anticipated that it would look like that and the overall shape was quite surprising,” said Guddat.
The study also revealed how the catalytic and regulatory subunits of the complex communicate with each other and provide a pathway for activation as well as to enable feedback inhibition by the three amino acids.
Identifying the location of the binding site on the subunit for valine and the way the structure changed had not been previously understood.
“Although there are only subtle changes to the structure, locking the protein into a specific single conformation is an important advance,” said Guddat
“Inhibiting the activity of the complex could lead to the discovery of new compounds to be used as improved herbicides or new drugs to treat serious invasive fungal infections,” said Guddat.
Another surprising finding of the research was finding a pocket in the core of the regulatory subunit which accommodates the molecule, ATP.
“It acts like a piece of glue to hold the structure together.”
The investigators reported that the highly ordered structure is conserved across three kingdoms of life. As well as the fungal and plant forms of the enzyme, the structure of AHAS was also determined from Mycobacterium tuberculosis.
Although progress had been made on parts of the structure in recent years, the team could not resolve the entire structure until they had prepared suitable samples for macromolecular crystallography and cryo-electron microscopy.
Both techniques are at the corner stone of modern structural biology.
Guddat collected approximately 50 data sets remotely on the macromolecular crystallography (MX) beamline at the Australian Synchrotron to obtain the best structure possible.
The plant enzyme is not highly soluble, which made it unsuitable for the production of crystals needed for the synchrotron.
Researchers at Shanghai Tech University assisted with cryo-electron microscopy to determine the structure of the plant enzyme.
“The higher resolution of the crystallographic images are sharper, clearer and more convincing. But, by using two methods, we were able to validate both structures against each other,” said Guddat.
Guddat thanked the macromolecular crystallography beamline scientists who assisted with the experiments over many months, including Dr Tom Caradoc-Davies and praised the reliability and speed at which data can be collected.
Dr Tom Caradoc-Davies, previously an MX beamline scientist who is now lead scientist for the new high performance macromolecular crystallography beamline (MX3) under development, congratulated Prof Guddat and his associates on resolving the structure and said it was an example of great scientific teamwork and showed the benefit of a fast detector and beamline.
“To be published in Nature indicates the calibre of this research and its importance,” said Caradoc-Davies.
“We have had a remote service operational for a number of years, well before the COVID epidemic,” he said.
The next goal for Guddat and associates is to provide a more detailed structure for AHAS from Mycobacterium tuberculosis, because there is a pressing need to find new treatments for drug resistant strains of this disease.
Lead author Dr Thierry Lonhienne carried out the majority of the study. PhD candidate, Yu Shang Low worked with the Shanghai Tech University in China to obtain the cryo-electron microscopy images.
Other collaborating institutions included ShanghaiTech University, Tsinghua University in China and Cambridge University, UK.
DOI: https://doi.org/10.1038/s41586-020-2514-3
Media report University of Queensland



